
Let’s improve on the emotion recognition from a previous article about FisherFace Classifiers. We will be using facial landmarks and a machine learning algorithm, and see how well we can predict emotions in different individuals, rather than on a single individual like in another article about theemotion recognising music player.
Important: The code in this tutorial is licensed under the GNU 3.0 open source license and you are free to modify and redistribute the code, given that you give others you share the code with the same right, and cite my name (use citation format below). You are not free to redistribute or modify the tutorial itself in any way. By reading on you agree to these terms. If you disagree, please navigate away from this page.
Citation format
van Gent, P. (2016). Emotion Recognition Using Facial Landmarks, Python, DLib and OpenCV. A tech blog about fun things with Python and embedded electronics. Retrieved from: http://www.paulvangent.com/2016/08/05/emotion-recognition-using-facial-landmarks/
IE users: I’ve gotten several reports that sometimes the code blocks don’t display correctly or at all on Internet Explorer. Please refresh the page and they should display fine.
Introduction and getting started
Using Facial Landmarks is another approach to detecting emotions, more robust and powerful than the earlier used fisherface classifier, but also requiring some more code and modules. Nothing insurmountable though. We need to do a few things:
- Get images from a webcam
- Detect Facial Landmarks
- Train a machine learning algorithm (we will use a linear SVM)
- Predict emotions
Those who followed the two previous posts about emotion recognition will know that the first step is already done.
Also we will be using:
- Python (2.7 or higher is fine, anaconda + jupyter notebook is a nice combo-package)
- OpenCV (I still use 2.4.9……so lazy, grab here)
- SKLearn (if you installed anaconda, it is already there, otherwise get it with pip install sklearn)
- Dlib (a C++ library for extracting the facial landmarks, see below for instructions)
- Visual Studio 2015 (get the community edition here, also select the Python Tools in the installation dialog).
- Note that VS is not strictly required, I just build the modules against it. However it is a very nice IDE that also has good Python bindings and allows you to quickly make GUI applications to wrap around your Python scripts. I would recommend you give it a go
Installing and building the required libraries
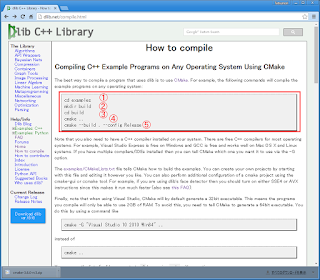
I am on Windows, and building libraries on Windows always gives many people a bad taste in their mouths. I can understand why, however it’s not all bad and often the problems people run into are either solved by correctly setting PATH variables, providing the right compiler or reading the error messages and installing the right dependencies. I will walk you through the process of compiling and installing Dlib.
First install CMake. This should be straightforward, download the windows installer and install. Make sure to select the option “Add CMake to the system PATH” during the install. Choose whether you want this for all users or just for your account.
Download Boost-Python and extract the package. I extracted it into C:\boost but it can be anything. Fire up a command prompt and navigate to the directory. Then do:
Once all is done you will find a folder named bin, or bin.v2, or something like this in your boost folder. Now it’s time to build Dlib.
Download Dlib and extract it somewhere. I used C:\Dlib but you can do it anywhere. Go back to your command prompt, or open a new one if you closed it, and navigate to your Dlib folder. Do this sequentially:
Open your Python interpreter and type “import dlib”. If you receive no messages, you’re good to go! Nice.
Testing the landmark detector
Before diving into much of the coding (which probably won’t be much because we’ll be recycling), let’s test the DLib installation on your webcam. For this you can use the following snippet. If you want to learn how this works, be sure to also compare it with the first script under “Detecting your face on the webcam” in the previous post. Much of the same OpenCV code to talk to your webcam, process the image by converting to grayscale, optimising the contrast with an adaptive histogram equalisation and displaying it is something we did there.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
This will result in your face with a lot of dots outlining the shape and all the “moveable parts”. The latter is of course important because it is what makes emotional expressions possible.



Note if you have no webcam and/or would rather like to try this on a static image, replace line #11 with something like frame = cv2.imread(“filename”) and comment out line #6 where we define the video_capture object. You will get something like:

my face has dots
people tell me my face has nice dots
experts tell me these are the best dots
I bet I have the best dots
Extracting features from the faces
The first thing to do is find ways to transform these nice dots overlaid on your face into features to feed the classifer. Features are little bits of information that describe the object or object state that we are trying to divide into categories. Is this description a bit abstract? Imagine you are in a room without windows with only a speaker and a microphone. I am outside this room and I need to make you guess whether there is a cat, dog or a horse in front of me. The rule is that I can only use visual characteristics of the animal, no names or comparisons. What do I tell you? Probably if the animal is big or small, that it has fur, that the fur is long or short, that it has claws or hooves, whether it has a tail made of flesh or just from hair, etcetera. Each bit of information I pass you can be considered afeature, and based the same feature set for each animal, you would be pretty accurate if I chose the features well.
How you extract features from your source data is actually where a lot of research is, it’s not just about creating better classifying algorithms but also about finding better ways to collect and describe data. The same classifying algorithm might function tremendously well or not at all depending on how well the information we feed it is able to discriminate between different objects or object states. If, for example, we would extract eye colour and number of freckles on each face, feed it to the classifier, and then expect it to be able to predict what emotion is expressed, we would not get far. However, the facial landmarks from the same image material describe the position of all the “moving parts” of the depicted face, the things you use to express an emotion. This is certainly useful information!
To get started, let’s take the code from the example above and change it so that it fits our current needs, like this:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
Here we extract the coordinates of all face landmarks. These coordinates are the first collection of features, and this might be the end of the road. You might also continue and try to derive other measures from this that will tell the classifier more about what is happening on the face. Whether this is necessary or not depends. For now let’s assume it is necessary, and look at ways to extract more information from what we have. Feature generation is always a good thing to try, if only because it brings you closer to the data and might give you ideas or alternative views at it because you’re getting your hands dirty. Later on we’ll see if it was really necessary at a classification level.
To start, look at the coordinates. They may change as my face moves to different parts of the frame. I could be expressing the same emotion in the top left of an image as in the bottom right of another image, but the resulting coordinate matrix would express different numerical ranges. However, the relationships between the coordinates will be similar in both matrices so some information is present in a location invariant form, meaning it is the same no matter where in the picture my face is.
Maybe the most straightforward way to remove numerical differences originating from faces in different places of the image would be normalising the coordinates between 0 and 1. This is easily done by:  , or to put it in code:
, or to put it in code:
However, there is a problem with this approach because it fits the entire face in a square with both axes ranging from 0 to 1. Imagine one face with its eyebrows up high and mouth open, the person could be surprised. Now imagine an angry face with eyebrows down and mouth closed. If we normalise the landmark points on both faces from 0-1 and put them next to each other we might see two very similar faces. Because both distinguishing features lie at the edges of the face, normalising will push both back into a very similar shape. The faces will end up looking very similar. Take a moment to appreciate what we have done; we have thrown away most of the variation that in the first place would have allowed us to tell the two emotions from each other! Probably this will not work. Of course some variation remains from the open mouth, but it would be better not to throw so much away.
A less destructive way could be to calculate the position of all points relative to each other. To do this we calculate the mean of both axes, which results in the point coordinates of the sort-of “centre of gravity” of all face landmarks. We can then get the position of all points relative to this central point. Let me show you what I mean. Here’s my face with landmarks overlaid:

First we add a “centre of gravity”, shown as a blue dot on the image below:

Lastly we draw a line between the centre point and each other facial landmark location:

Note that each line has both a magnitude (distance between both points) and a direction (angle relative to image where horizontal=0°), in other words, a vector.
But, you may ask, why don’t we take for example the tip of the nose as the central point? This would work as well, but would also throw extra variance in the mix due to short, long, high- or low-tipped noses. The “centre point method” also introduces extra variance; the centre of gravity shifts when the head turns away from the camera, but I think this is less than when using the nose-tip method because most faces more or less face the camera in our sets. There are techniques to estimate head pose and then correct for it, but that is beyond this article.
There is one last thing to note. Faces may be tilted, which might confuse the classifier. We can correct for this rotation by assuming that the bridge of the nose in most people is more or less straight, and offset all calculated angles by the angle of the nose bridge. This rotates the entire vector array so that tilted faces become similar to non-tilted faces with the same expression. Below are two images, the left one illustrates what happens in the code when the angles are calculated, the right one shows how we can calculate the face offset correction by taking the tip of the nose and finding the angle the nose makes relative to the image, and thus find the angular offset β we need to apply.

Now let’s look at how to implement what I described above in Python. It’s actually fairly straightforward. We just slightly modify the get_landmarks() function from above.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
That was actually quite manageable, no? Now it’s time to put all of the above together with some stuff from the first post. The goal is to read the existing dataset into a training and prediction set with corresponding labels, train the classifier (we use Support Vector Machines with linear kernel from SKLearn, but feel free to experiment with other available kernels such as polynomial or rbf, or other classifiers!), and evaluate the result. This evaluation will be done in two steps; first we get an overall accuracy after ten different data segmentation, training and prediction runs, second we will evaluate the predictive probabilities.
Déja-Vu All Over Again
The next thing we will be doing is returning to the two datasets from the original post. Let’s see how this approach stacks up.
First let’s write some code. The approach is to first extract facial landmark points from the images, randomly divide 80% of the data into a training set and 20% into a test set, then feed these into the classifier and train it on the training set. Finally we evaluate the resulting model by predicting what is in the test set to see how the model handles the unknown data. Basically a lot of the steps are the same as what we did earlier.
The quick and dirty (I will clean and ‘pythonify’ the code later, when there is time) solution based off of earlier code could be something like:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
Remember that in the previous post, for the standard set at 8 categories we managed to get 69.3% accuracy with the FisherFace classifier. This approach yields 84.1% on the same data, a lot better!
We then reduced the set to 5 emotions (leaving out contempt, fear and sadness), because the 3 categories had very few images, and got 82.5% correct. This approach gives 92.6%, also much improvement.
After adding the less standardised and more difficult images from google, we got 61.6% correct when predicting 7 emotions (the contempt category remained very small so we left that out). This is now 78.2%, also quite an improvement. This remains the lowest accuracy, showing that for a more diverse dataset the problem is also more difficult. Keep in mind that the dataset I use is still quite small in machine learning terms, containing about 1000 images spread over 8 categories.
Looking at features
So we derived different features from the data, but weren’t sure whether this was strictly necessary. So was this necessary? It depends! It depends on if doing so adds more unique variance related to what you’re trying to predict, it depends on what classifier you use, etc.
Let’s run different feature combinations as inputs through different classifiers and see what happens. I’ve run all iterations on the same slice of data with 4 emotion categories of comparable size (so that running the same settings again yields the same predictive value).
Using all of the features described so far leads to:
Linear SVM: 93.9%
Polynomial SVM: 83.7%
Random Forest Classifier: 87.8%
Now using just the vector length and angle:
Linear SVM: 87.8%
Polynomial SVM: 87.8%
Random Forest Classifier: 79.6%
Now using just the raw coordinates:
Linear SVM: 91.8%
Polynomial SVM: 89.8%
Random Forest Classifier: 59.2%
Now replacing all training data with zeros:
Linear SVM: 32.7%
Polynomial SVM: 32.7%
Random Forest Classifier: 32.7%
Now this is interesting! First note that there isn’t much difference in the accuracy of the support vector machine classifiers when using the extra features we generate. This type of classifier already preprocesses the data quite extensively. The extra data we generate does not contain much if any extra information to this classifier, so it only marginally improves the performance of the linear kernel, and actually hurts the polynomial kernel because data with a lot of overlapping variance can also make a classification task more difficult. By the way, this is a nice 2D visualisation of what an SVC tries to achieve, complexity escalates when adding one dimension. Now remember that the SVC operates in an N-dimensional space and try to imagine what a set of hyperplanes in 4, 8, 12, 36 or more dimensions would look like. Don’t drive yourself crazy.
Random Forest Classifiers do things a lot differently. Essentially they are a forest of decision trees. Simplified, each tree is a long list of yes/no questions, and answering all questions leads to a conclusion. In the forest the correlation between each tree is kept as low as possible, which ensures every tree brings something unique to the table when explaining patterns in the data. Each tree then votes on what it thinks the answer is, and most votes win. This approach benefits extensively from the new features we generated, jumping from 59.2% to 87.8% accuracy as we combine all derived features with the raw coordinates.
So you see, the answer you likely get when you ask any scientist a direct question holds true here as well: it depends. Check your data, think twice and try a few things.
The last that may be noticed is that, when not adding any data at all and in stead presenting the classifiers with a matrix of zeros, they still perform slightly above the expected chance level of 25%. This is because the categories are not identically sized.
Looking at mistakes
Lastly, let’s take a look at where the model goes wrong. Often this is where you can learn a lot, for example this is where you might find that a single category just doesn’t work at all, which can lead you to look critically at the training material again.
One advantage of the SVM classifier we use is that it is probabilistic. This means that it assigns probabilities to each category it has been trained for (and you can get these probabilities if you set the ‘probability’ flag to True). So, for example, a single image might be “happy” with 85% probability, “angry” with “10% probability, etc.
To get the classifier to return these things you can use its predict_proba() function. You give this function either a single data row to predict or feed it your entire dataset. It will return a matrix where each row corresponds to one prediction, and each column represents a category. I wrote these probabilities to a table and included the source image and label. Looking at some mistakes, here are some notable things that were classified incorrectly (note there are only images from my google set, the CK+ set’s terms prohibit me from publishing images for privacy reasons):

anger: 0.03239878
contempt: 0.13635423
disgust: 0.0117559
fear: 0.00202098
neutral: 0.7560004
happy: 0.00382895
sadness: 0.04207027
surprise: 0.0155695
The correct answer is contempt. To be honest I would agree with the classifier, because the expression really is subtle. Note that contempt is the second most likely according to the classifier.

anger: 0.0726657
contempt: 0.24655082
disgust: 0.06427896
fear: 0.02427595
neutral: 0.20176133
happy: 0.03169822
sadness: 0.34911036
surprise: 0.00965867
The correct answer is disgust. Again I can definitely understand the mistake the classifier makes here (I might make the same mistake..). Disgust would be my second guess, but not the classifier’s. I have removed this image from the dataset because it can be ambiguous.

anger: 0.00304093
contempt: 0.01715202
disgust: 0.74954754
fear: 0.04916257
neutral: 0.00806644
happy: 0.13546932
sadness: 0.02680473
surprise: 0.01075646
The correct answer is obviously happy. This is a mistake that is less understandable but still the model is quite sure (~75%). There definitely is no hint of disgust in her face. Do note however, that happiness would be the classifier’s second guess. More training material might rectify this situation.

anger: 0.0372873
contempt: 0.08705531
disgust: 0.12282577
fear: 0.16857784
neutral: 0.09523397
happy: 0.26552763
sadness: 0.20521671
surprise: 0.01827547
The correct answer is sadness. Here the classifier is not sure at all (~27%)! Like in the previous image, the second guess (~20%) is the correct answer. This may very well be fixed by having more (and more diverse) training data.

anger: 0.01440529
contempt: 0.15626157
disgust: 0.01007962
fear: 0.00466321
neutral: 0.378776
happy: 0.00554828
sadness: 0.07485257
surprise: 0.35541345
The correct answer is surprise. Again a near miss (~38% vs ~36%)! Also note that this is particularly difficult because there are few baby faces in the dataset. When I said earlier that the extra google images are very challenging for a classifier, I meant it!
Upping the game – the ultimate challenge
Although the small google dataset I put together is more challenging than the lab-conditions of the CK/CK+ dataset, it is still somewhat controlled. For example I filtered out faces that were more sideways than frontal-facing, where the emotion was very mixed (happily surprised for example), and also where the emotion was so subtle that even I had trouble identifying it.
A far greater (and more realistic still) challenge is the SFEW/AFEW dataset, put together from a large collection of movie scenes. Read more about it here. The set is not publicly available but the author was generous enough to share the set with me so that I could evaluate the taken approach further.
Guess what, it fails miserably! It attained about 44.2% on the images when training on 90% and validating on 10% of the set. Although this is on par with what is mentioned in the paper, it shows there is still a long way to go before computers can recognize emotions with a high enough accuracy in real-life settings. There are also video clips included on which we will spend another post together with convolutional neural nets at a later time.
This set is particularly difficult because it contains different expressions and facial poses and rotations for similar emotions. This was the purpose of the authors; techniques by now are good enough to recognise emotions on controlled datasets with images taken in lab-like conditions, approaching upper 90% accuracy in many recent works (even our relatively simple approach reached early 90). However these sets do not represent real life settings very much, except maybe when using laptop webcams, because you always more or less face this device and sit at a comparable distance when using the laptop. This means for applications in marketing and similar fields the technology is already usable, albeit with much room for improvement still available and requiring some expertise to implement it correctly.
Final reflections
Before concluding I want you to take a moment, relax and sit back and think. Take for example the SFEW set with real-life examples, accurate classification of which quickly gets terribly difficult. We humans perform this recognition task remarkably well thanks to our highly complex visual system, which has zero problems with object rotation in all planes, different face sizes, different facial characteristics, extreme changes in lighting conditions or even partial occlusion of a face. Your first response might be “but that’s easy, I do it all the time!”, but it’s really, really, really not. Think for a moment about what an enormously complex problem this really is. I can show you a mouth and you would already be quite good at seeing an emotion. I can show you about 5% of a car and you could recognize it as a car easily, I can even warp and destroy the image and your brain would laugh at me and tell me “easy, that’s a car bro”. This is a task that you solve constantly and in real-time, without conscious effort, with virtually 100% accuracy and while only using the equivalent of ~20 watts for yourentire brain (not just the visual system). The average still-not-so-good-at-object-recognition CPU+GPU home computer uses 350-450 watts when computing. Then there’s supercomputers like theTaihuLight, which require about 15.300.000 watts (using in one hour what the average Dutch household uses in 5.1 years). At least at visual tasks, you still outperform these things by quite a large margin with only 0.00013% of their energy budget. Well done, brain!
Anyway, to try and tackle this problem digitally we need another approach. In another post we will look at various forms of neural nets (modeled after your brain) and how these may or may not solve the problem, and also at some other feature extraction techniques.
The CK+ dataset was used for validating and training of the classifier in this article, references to the set are:
- Kanade, T., Cohn, J. F., & Tian, Y. (2000). Comprehensive database for facial expression analysis. Proceedings of the Fourth IEEE International Conference on Automatic Face and Gesture Recognition (FG’00), Grenoble, France, 46-53.
- Lucey, P., Cohn, J. F., Kanade, T., Saragih, J., Ambadar, Z., & Matthews, I. (2010). The Extended Cohn-Kanade Dataset (CK+): A complete expression dataset for action unit and emotion-specified expression. Proceedings of the Third International Workshop on CVPR for Human Communicative Behavior Analysis (CVPR4HB 2010), San Francisco, USA, 94-101.
The SFEW/AFEW dataset used for evaluation is authored by and described in:
- A. Dhall, R. Goecke, S. Lucey and T. Gedeon, “Collecting Large, Richly Annotated Facial- Expression Databases from Movies”, IEEE MultiMedia 19 (2012) 34-41.
- A. Dhall, R. Goecke, J. Joshi, K. Sikka and T. Gedeon, “ Emotion Recognition In The Wild Challenge 2014: Baseline, Data and Protocol”, ACM ICMI 2014.





















 Universal-USB-Installer-1.9.5.5.exe
Universal-USB-Installer-1.9.5.5.exe





















































 http_kaylab.tistory.com_1.pdf
http_kaylab.tistory.com_1.pdf
































cmake . -DBoost_DEBUG=ON? – Fraser Jun 11 '14 at 22:03