기계 학습(Machine Learning)은 즐겁다! Part 2
기계 학습을 사용해서 Super Mario Maker의 레벨 제작하기
Part 1에서는 기계 학습(Machine Learning)이 문제를 해결하기 위해 코드를 전혀 작성하지 않고도, 일반 알고리즘(generic algorithm)을 사용해서 주어진 데이터에서 흥미로운 것을 알아 낼 수 있다는 것을 알아봤습니다. (아직 Part 1을 읽지 않았다면, 지금 읽어보세요!)
이번에는 이러한 일반 알고리즘 중 하나로 아주 멋진 일을 해내는 것을 보게 될텐데 – 바로 사람들이 만든 것처럼 보이는 비디오 게임의 레벨을 제작하는 것입니다. 우리는 신경망(neural network)를 만들고 기존의 슈퍼 마리오 레벨들을 통해서 새로운 슈퍼 마리오 레벨이 쉽게 만들어지는 것을 살펴 볼 예정입니다.
Part 1과 마찬가지로, 이 안내서는 기계 학습에 궁금한 점은 있지만 어디서부터 시작해야할지 모르는 사람들을ㅍ위한 것입니다. 이 글의 목표는 누구에게나 쉽게 다가가는 데 있습니다 – 이는 글에 많은 일반화가 있음을 의미합니다. 하지만 어떻습니까? 그래서 더 많은 사람들이 ML에 관심을 가지게 된다면, 목표를 달성한 것입니다.
좀더 영리하게 추측하기
Part 1에서, 우리는 주택의 속성을 기반으로 주택 가치를 추정하는 간단한 알고리즘을 만들었습니다. 어떤 주택에 대한 데이터가 다음과 같다고 가정해 봅시다:

우리는 간단한 추정 함수(estimation function)를 만들었습니다:
def estimate_house_sales_price(num_of_bedrooms, sqft, neighborhood):
price = 0
# 이건 한 꼬집 넣고
price += num_of_bedrooms * 0.123
# 그리고 저건 한 스픈 정도 넣고
price += sqft * 0.41
# 이건 아마도 한 줌 넣고
price += neighborhood * 0.57
return price
즉, 우리는 각 속성에 가중치를 곱하여 주택 가격을 추정했습니다. 그런 다음 이 값들을 더해서 주택의 최종 가치를 얻었습니다.
코드를 사용하는 대신, 간단한 다이어그램으로 해당 함수를 표현해 보겠습니다:
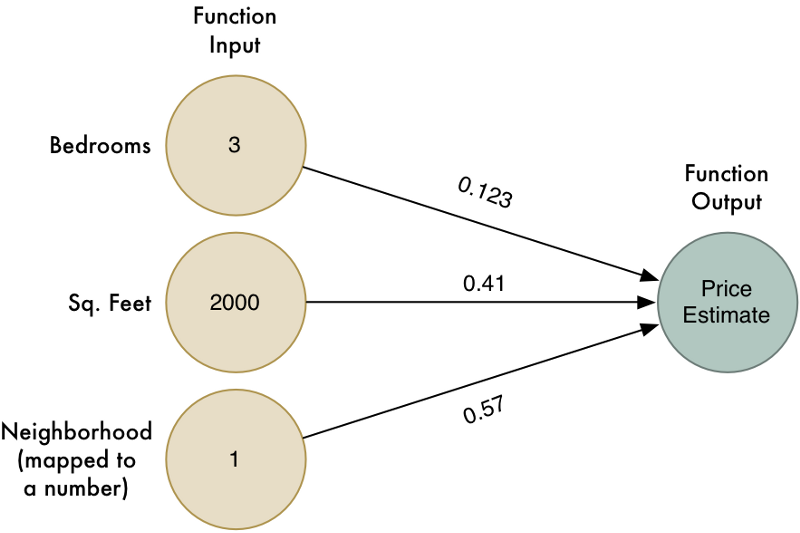
하지만, 이 알고리즘은 입력데이터와 결과 사이에 선형(linear) 관계가 있는 단순한 문제에 대해서만 동작합니다. 주택 가격이 실제로 그렇게 단순하지 않다면 어떻게 될까요? 예를 들어, 큰 주택과 작은 주택에서는 이웃이 크게 중요 할 수 있지만 중간 크기의 집에서는 중요하지 않을 수 있습니다. 우리 모델에 이런 종류의 복잡한 세부 사항을 적용할 수 있을까요?
더 영리해지기 위해서, 우리는 각각 다른 경우에 적용되는 서로 다른 가중치를 사용해서 알고리즘을 여러 번 실행해 볼 수 있습니다:
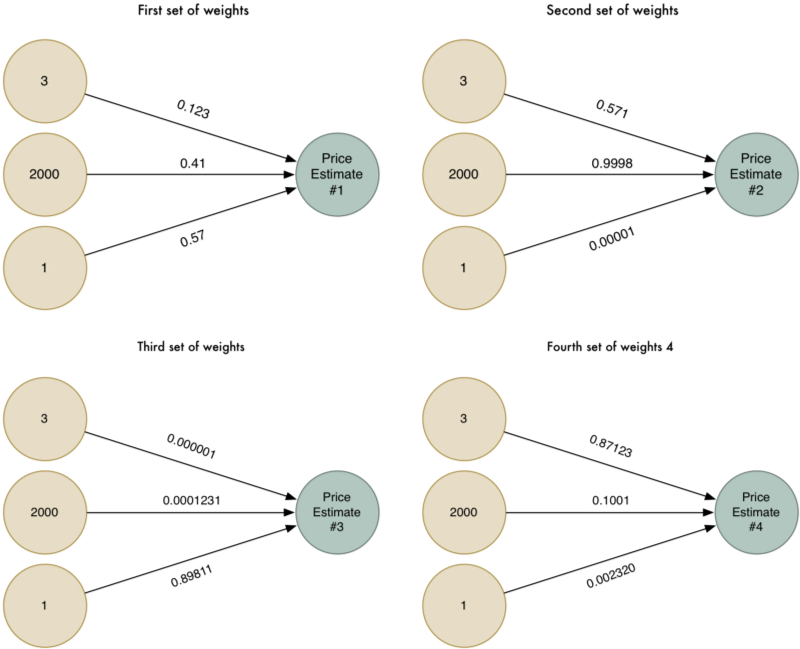
이제 서로 다른 네가지의 가격 예측이 있습니다. 이 네 가지 가격 예측들을 하나의 최종 예측으로 결합해 보겠습니다. 동일한 알고리즘으로 다시 실행할 것입니다 (다만, 다른 가중치의 세트 사용해서)!
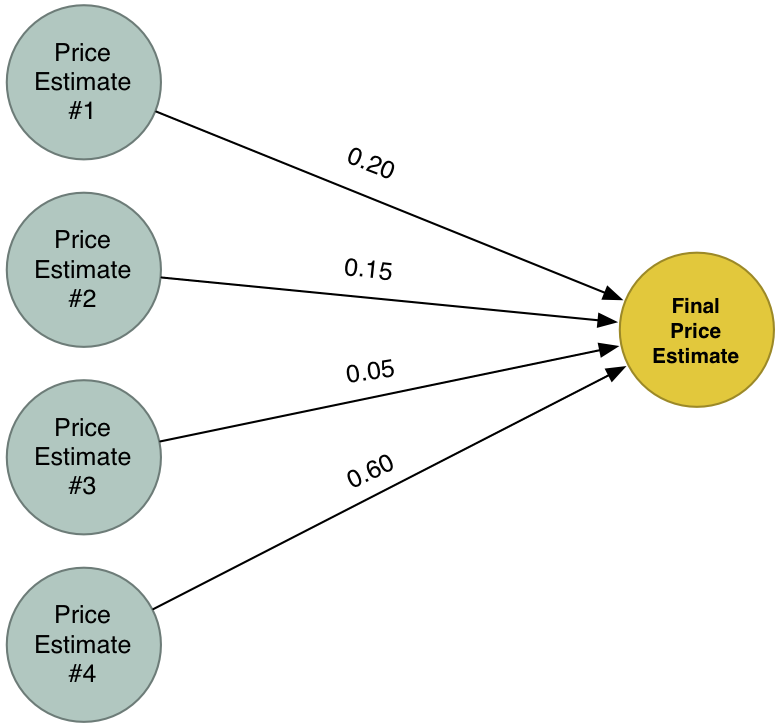
우리의 새로운 최종 해답은 문제를 해결하기위한 네 가지 시도의 예측들을 결합한 것입니다. 이러한 방법을 이용하면, 하나의 간단한 모델에서 다룰 수 있는 것보다 더 많은 사례에 대한 모델링 할 수 있습니다.
신경망(Neural Network)이란 무엇인가요?
우리의 네가지 시도를 하나의 큰 다이어그램으로 결합해 보겠습니다.
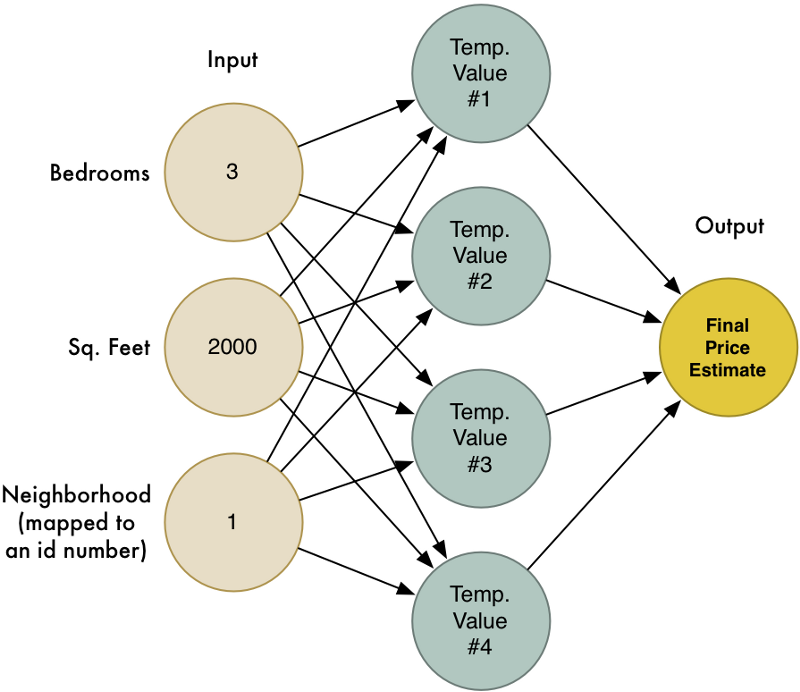
이것이 바로 신경망(neural network)입니다! 각 노드는 일련의 입력을 받아 들여 가중치를 적용하고, 출력 값을 계산하는 방법을 알고 있습니다. 이렇게 많은 노드를 서로 연결함으로써, 우리는 복잡한 함수를 모델링 할 수 있습니다.
이 글을 쉽게 유지하기 위해서 (feature scaling과 activation function을 포함해서) 많은 부분을 건너 뛰었지만, 여기서 가장 중요한 것은 다음의 기본 아이디어입니다.
- 우리는 일련의 입력을 받고 가중치를 곱해 출력을 얻는 간단한 추정 함수를 만들었습니다. 이 간단한 함수를 뉴런(neuron)이라고 부르겠습니다.
- 단순한 뉴런들(neurons)을 서로 연결함으로써, 우리는 하나의 단일 뉴런으로 모델링하기에는 너무 복잡한 함수를 모델링 할 수 있습니다.
이것은 LEGO와 같습니다! 하나의 LEGO 블록으로는 많은 모델을 만들 수는 없지만, 조립할 수 있는 기본 LEGO 블록이 충분하다면 어떤 것이라도 모델링 할 수 있습니다.

신경망(Neural Network)에 메모리 주기
앞서 우리가 본 신경망은 동일한 입력을 주면 항상 같은 결과을 반환합니다. 메모리(기억 장치)가 없기 때문입니다. 프로그래밍 용어로 말하면, 이것은 상태 비저장 알고리즘(stateless algorithm)입니다.
(집값 추정과 같은) 많은 경우에, 이 방식은 당신이 정확히 원하는 것입니다. 그러나 이런 종류의 모델이 할 수없는 한 가지는 시간이 지남에 따라 변하는 데이터의 패턴에 대응하는 것입니다.
제가 키보드를 건네주고 당신에게 이야기를 하나 써보라고 하는 상황을 상상해보십시오. 그런데 시작하기 전에, 내 직업은 사람들이 타이핑 할 첫 번째 글자를 예측하는 것입니다. 어떤 글자를 예측해야 할까요?
저는 제 영어 지식을 사용해서 올바른 글자를 예측하는 확률을 높일 수 있습니다. 예를 들어, 당신은 단어의 첫글자에 많이 사용되는 글자를 입력할 지도 모릅니다. 만약 당신이 과거에 쓴 이야기를 봤다면, 아마도 이야기의 시작 부분에서 일반적으로 사용하는 단어를 바탕으로 좀더 좁힐 수도 있을 것입니다. 일단 모든 데이터가 확보되면, 신경망을 구축해서 당신이 글을 시작할 때 특정 글자를 얼마나 사용할 가능성이 있는지를 모델링할 수 있을 것입니다.
우리는 모델은 아마 다음과 같을 것입니다:
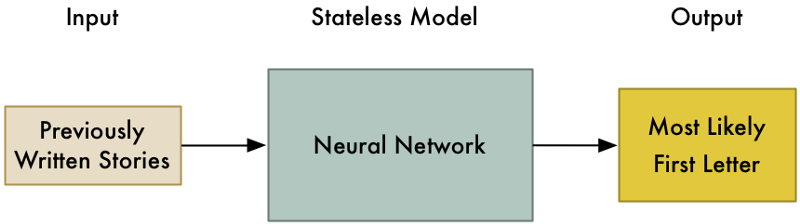
이제 문제를 좀 더 어렵게 만들어 보겠습니다. 당신의 이야기의 어느 부분에서든 입력 할 다음 글자를 추측해야 한다고 가정해 봅시다. 훨씬 더 흥미로운 문제가 되었습니다.
어니스트 헤밍웨이(Ernest Hemingway)의 The Sun Also Rises의 처음 몇 단어를 예로 들어 보겠습니다.
Robert Cohn was once middleweight boxi
자 다음에 올 글자는 무엇일까요?
당신은 ‘n’을 추측했을 것입니다 — 그 단어는 아마도 boxing 일 것입니다. 우리는 저 문장에서 보이는 글자들과 영어의 일반적인 단어에 대한 지식을 바탕으로 이를 알 수 있습니다. 또한, ‘미들급’(‘middleweight’)이라는 단어는 권투에 대해 이야기하고 있다는 추가 단서를 제공해 줍니다.
신경망으로이 문제를 해결하려면, 우리 모델에 상태(state)를 추가해야합니다. 우리가 신경망에 응답을 요청할 때마다 우리는 중간 계산 결과들을 저장하고 다음 입력의 일부로써 다시 사용할 수 있습니다. 그렇게 하면, 우리 모델은 최근의 입력을 기반으로 예측을 조정하게 됩니다.
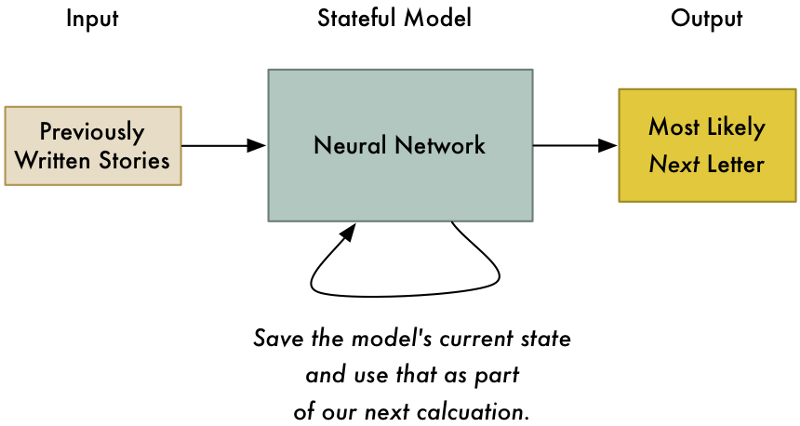
우리 모델에서 상태를 기록해 두면 단지 그 이야기에서 가장 가능성있는 첫 글자를 예측하는 것이 아니라 이전의 모든 문자를 고려해서 가장 가능성있는 다음 글자를 예측할 수 있게 됩니다.
글자 하나 예측하는게 무슨 소용이 있나요?
이야기 안에서 다음 글자를 예측하는 것은 꽤 쓸모없는 것처럼 보일 수 있습니다. 여기서 중요한 것은 무엇인가요?
멋진 예 중에 하나가 바로 휴대 전화 키보드에 있는 자동 예측(auto-predict) 기능입니다:
이제 이 아이디어를 극단적 끌고 가면 어떻게 될까요? 모델에 다음으로 가장 가능성있는 글자를 계속해서 아니 영원히 예측 해달라고 요청하면 어떻게 될까요? 차라리 완전한 이야기를 하나 만들어 달라고 요청하는 편이 낫겠네요!
이야기 만들기
우리는 헤밍웨이의 다음 문장을 어떻게 추측 할 수 있는지 알아보았습니다. 이제 헤밍웨이 스타일로 전체 이야기를 만들어 봅니다.
이를 위해, 우리는 Andrej Karpathy가 작성한 순한 신경망 구현(Recurrent Neural Network implementation)을 사용할 것입니다. Andrej는 Stanford의 Deep-Learning 연구원이며 RNN을 사용하여 텍스트를 생성에 대해 훌륭한 글을 썼습니다. github에서 이 모델에 관한 모든 코드를 볼 수 있습니다.
84개의 고유한 문자(구두점, 대문자/소문자 등 포함)를 사용해서 총 362,239 글자로 쓰여진 The Sun Also Rises 의 전체 텍스트로부터 모델을 제작할 것입니다. 이 데이터 세트는 현실에서 사용되는 전형적인 애플리케이션에 비하면 아주 적은 양입니다. 헤밍웨이의 스타일의 정말 좋은 모델을 제작하려면, 여러 번에 걸처 많은 샘플 텍스트를 사용하는 편이 훨씬 낫습니다. 뭐 이정도면 예를 들어 해보는데는 그런데로 충분해 보입니다.
이제 막 RNN을 훈련시키기 시작했기 때문에, 바로 글자를 예측하는 것은 그리 좋지 않습니다. 훈련을 100 회 반복 한 후 작성된 내용은 다음과 같습니다:
hjCTCnhoofeoxelif edElobe negnk e iohehasenoldndAmdaI ayio pe e h’e btentmuhgehi bcgdltt. gey heho grpiahe.
Ddelnss.eelaishaner” cot AAfhB ht ltny
ehbih a”on bhnte ectrsnae abeahngy
amo k ns aeo?cdse nh a taei.rairrhelardr er deffijha
단어들 사이에 가끔씩 공백이 있다는 정도는 알아냈지만, 이게 전부입니다.
1000 회 정도 반복 한 후에는 가능성이 있어 보입니다:
hing soor ither. And the caraos, and the crowebel for figttier and ale the room of me? Streat was not to him Bill-stook of the momansbed mig out ust on the bull, out here. I been soms
inick stalling that aid.
“Hon’t me and acrained on .Hw’s don’t you for the roed,” In’s pair.”
“Alough marith him.”
모델은 이제 기본 문장 구조의 패턴을 식별하기 시작했습니다. 문장의 끝 부분에 마침표를 찍고 심지어 묻고 답하는 대화도 추가했습니다. 몇 마디는 알아볼 수 있지만, 여전히 많은 부분이 말도 안됩니다.
그러나 수천 번의 훈련을 수행하니 꽤 괜찮아 보입니다:
He went over to the gate of the café. It was like a country bed.
“Do you know it’s been me.”
“Damned us,” Bill said.
“I was dangerous,” I said. “You were she did it and think I would a fine cape you,” I said.
“I can’t look strange in the cab.”
“You know I was this is though,” Brett said.
“It’s a fights no matter?”
“It makes to do it.”
“You make it?”
“Sit down,” I said. “I wish I wasn’t do a little with the man.”
“You found it.”
“I don’t know.”
“You see, I’m sorry of chatches,” Bill said. “You think it’s a friend off back and make you really drunk.”
이 시점에서, 알고리즘은 헤밍웨이의 짧고 직접적인 대화 방식의 기본 패턴을 감지해 냈습니다. 몇가지 문장은 대충 말이 됩니다.
실제 책에 있는 텍스트와 비교해 보겠습니다:
There were a few people inside at the bar, and outside, alone, sat Harvey Stone. He had a pile of saucers in front of him, and he needed a shave.
“Sit down,” said Harvey, “I’ve been looking for you.”
“What’s the matter?”
“Nothing. Just looking for you.”
“Been out to the races?”
“No. Not since Sunday.”
“What do you hear from the States?”
“Nothing. Absolutely nothing.”
“What’s the matter?”
한 번에 한 글자를 예측하는 패턴을 찾는 것만으로도, 우리의 알고리즘은 올바른 형식으로 그럴듯하게 보이는 산문을 재현해 냈습니다. 정말 놀라운 일입니다!
텍스트를 처음부터 완전하게 만들 필요는 없습니다. 그냥 알고리즘에 처음 몇 글자를 제공하고 스스로 다음 몇 글자 찾도록 내버려 두면 됩니다.
재미삼아, 상상의 책을 위한 가짜 책표지를 만들어 보겠습니다. 이를 위해서 “Er”, “He”및 “The S”을 시드(seed)로 해서 새로운 저자와 책제목을 만들어 보겠습니다.

뭐 그런데로 괜찮네요!
그런데 정말로 놀라운 것은 이 알고리즘이 모든 일련의 데이터에서 패턴을 파악할 수 있다는 점입니다. 진짜처럼 보이는 조리법이나 가짜 오바마 연설문을 쉽게 만들 수 있습니다. 그렇다면 인간의 언어에만 제한을 둘 필요가 있을까요? 아닙니다, 우리는 패턴이있는 모든 종류의 데이터에 이 아이디어를 적용 할 수 있습니다.
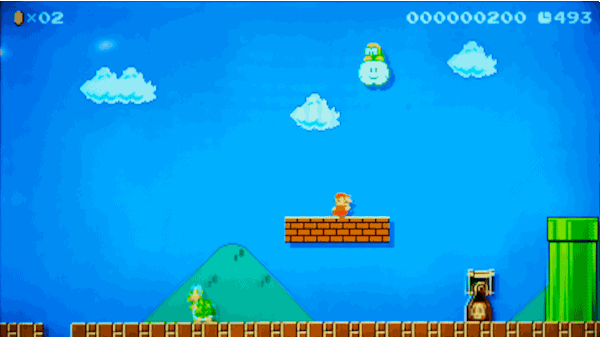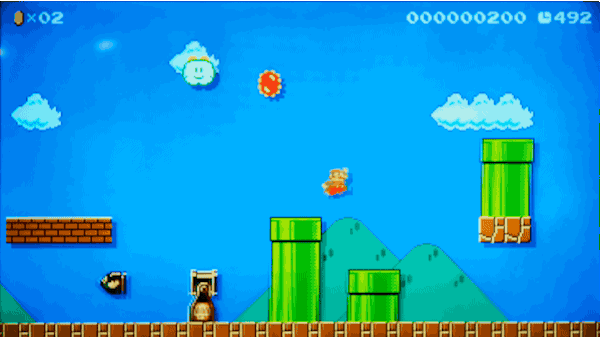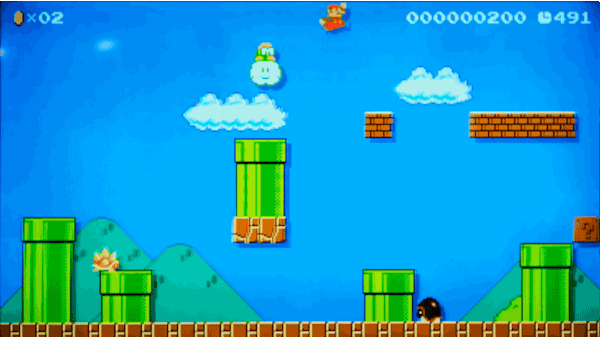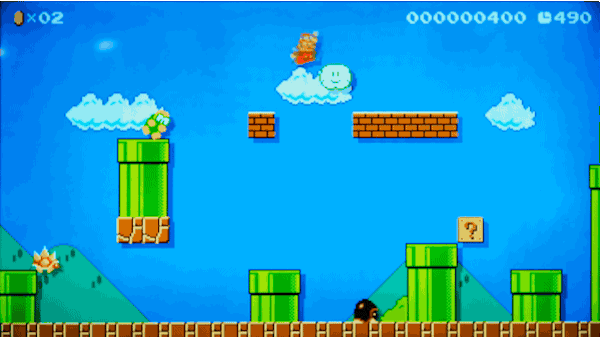
실제로 마리오 게임 레벨을 만들지 않고 마리오 게임 레벨 만들기
2015년, Nintendo사는 Wii U 게임 시스템 용 Super Mario Maker™를 출시했습니다.
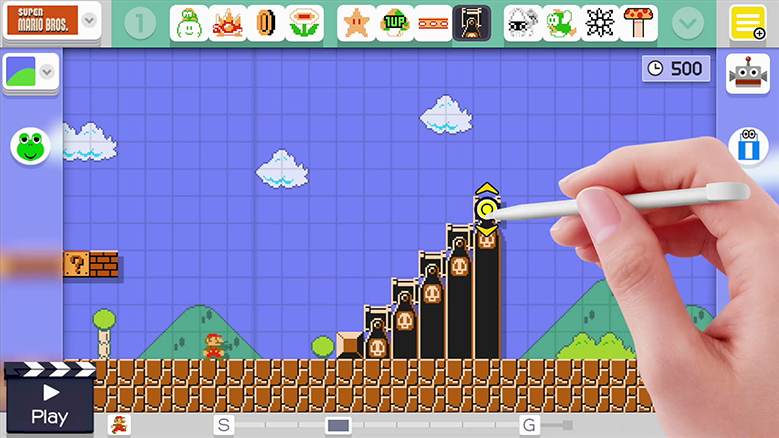
이 게임을 사용하면 게임패드에서 자신만의 슈퍼 마리오 브라더스(Super Mario Brothers) 레벨을 그릴 수 있고, 이를 인터넷에 업로드하면 친구들이 이 레벨을 플레이할 수도 있습니다. 자신만의 레벨에 실제 마리오 게임의 모든 기존 파워업(power-ups)과 적을 집어 넣을 수 있습니다. 이는 마치 슈퍼 마리오 브라더스를 플레이하면서 자란 사람들 위한 가상의 레고 세트와 같습니다.
가짜로 헤밍웨이의 글을 만들어낸 동일한 모델을 사용해서 가짜 슈퍼 마리오 브라더스 레벨을 만들어 낼 수 있을까요?
우선, 우리의 모델을 훈련시키 데이터 세트가 필요합니다. 1985 년에 출시 된 진짜 슈퍼 마리오 브라더스 게임의 모든 야외 레벨(outdoor levels)을 사용하겠습니다.

이 게임에는 32개의 레벨을 있으며, 그중 대략 70%가 동일한 야외 스타일(outdoor style) 입니다. 그래서 우리는 여기에 집중하겠습니다.
각 레벨의 디자인을 얻기위해서, 게임 원본의 게임 메모리에서 레벨 디자인을 빼내는 프로그램을 작성했습니다. 슈퍼 마리오 브라더스(Super Mario Bros.)는 30년이나 된 게임이라, 레벨이 게임의 메모리에 어떻게 저장되는 지 알 수 있는 온라인 리소스가 많이 있습니다. 오래된 비디오 게임에서 레벨 데이터를 추출하는 것은 언젠가 시도해 봐야하는 재미있는 프로그래밍 연습입니다.
게임에서 추출한 첫 번째 레벨입니다 (이 게임을 해본적이 있다면 아마도 기억할 것입니다):

자세히 살펴보면 레벨은 단순한 격자 객체들로 구성되어 있음을 알 수 있습니다:
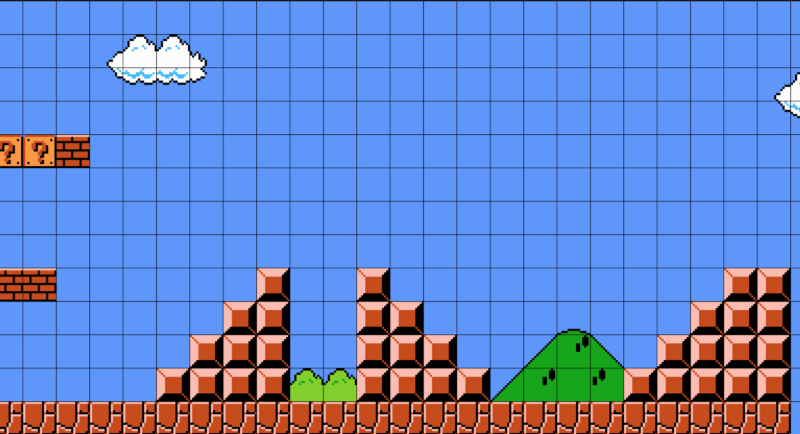
우리는 각 객체를 표현하는 문자를 사용해서 이 격자를 일련의 문자열로 쉽게 나타낼 수 있습니다.
--------------------------
--------------------------
--------------------------
#??#----------------------
--------------------------
--------------------------
--------------------------
-##------=--=----------==-
--------==--==--------===-
-------===--===------====-
------====--====----=====-
=========================-
우리는 레벨에 있는 각 객체를 문자로 대체했습니다:
- ‘-’ 는 빈공간
- ‘=’ 는 딱딱한 블럭
- ‘#’ 는 깰 수 있는 벽돌
- ‘?’ 는 코인 블럭
… 등등, 레벨에 있는 여러 객체들에 각각 다른 문자를 사용했습니다.
결과적으로 다음과 같이 텍스트 파일이 만들어 졌습니다.

이 텍스트 파일을 살펴보면, 마리오의 레벨은 줄(line) 단위로 보면 실제로 별다른 패턴이 없다는 것을 알 수 있습니다

줄 단위로 읽어 보면 실제로 캡처할 패턴이 없습니다. 많은 줄들이 완전히 비어 있습니다.
레벨의 패턴은 레벨을 일련의 열(column)으로 생각할 때 드디어 들어나게 됩니다.

열 단위로 보면 실제 패턴이 있습니다. 예를 들어, 각 열은 ‘=’로 끝납니다.
따라서 알고리즘이 데이터에서 패턴을 찾을 수 있게 하기 위해서는 데이터를 열별로 제공(feed)해야합니다. 입력 데이터의 가장 효과적인 표현을 찾는 것(feature selection이라고 함)은 기계 학습 알고리즘을 잘 사용하기 위한 중요한 요소 중에 하나입니다.
모델을 훈련시키기 위해서, 텍스트 파일을 90도 회전해야 했습니다. 이를 통해 패턴이 보다 쉽게 들어나는 순서에 따라 모델에 문자열을 제공(feed)할 수 있었습니다.
-----------=
-------#---=
-------#---=
-------?---=
-------#---=
-----------=
-----------=
----------@=
----------@=
-----------=
-----------=
-----------=
---------PP=
---------PP=
----------==
---------===
--------====
-------=====
------======
-----=======
---=========
---=========
모델 훈련시키기
헤밍웨이의 산문을 위한 모델을 만들 때 확인했듯이, 훈련시킬 수록 모델은 향상됩니다.
약간의 훈련 후, 우리 모델은 아직 쓸모없는 것을 만들고 있습니다:
--------------------------
LL+<&=------P-------------
--------
---------------------T--#--
-----
-=--=-=------------=-&--T--------------
--------------------
--=------$-=#-=-_
--------------=----=<----
-------b
-
현재는 ‘-’와 ‘=’가 많이 나타나야 한다와 같은 생각을 가지고 있지만, 그 뿐입니다. 아직 패턴을 전혀 알아 내지 못했습니다
수천 번의 반복을 진행하자, 이제 다음과 같은 걸 보기 시작합니다:
--
-----------=
----------=
--------PP=
--------PP=
-----------=
-----------=
-----------=
-------?---=
-----------=
-----------=
이제 모델은 각 줄이 동일한 길이여야한다는 것을 거의 알아 냈습니다. Mario의 로직 일부를 이해하기 시작했습니다: 마리오에서 파이프는 항상 2 블럭 넓이이고 높이는 최소 2 블럭 이상이므로 데이터에 “P”들은 2x2 클러스터로 나타나야 합니다. 굉장히 멋지군요!
*역자주: 여기서 2x2 클러스터란 “P”는 2글자 이상이 붙어서 두 줄에 걸처 연결된 형태(cluster)로 나타나야 한다는 의미
더 많은 훈련을 하면, 모델은 결국 완벽하게 유효한 데이터를 생성하게 됩니다:
--------PP=
--------PP=
----------=
----------=
----------=
---PPP=---=
---PPP=---=
----------=
모델에서 만들어진 전체 레벨의 가치 있는 데이터를 가져와 수평으로 회전시켜 보겠습니다:

이 데이터는 정말 멋집니다! 그리고 대단한 것들이 몇 가지 있습니다.
- 진짜 마리오 레벨처럼 Lakitu(구름을 타고 떠다니는 괴물)를 레벨의 시작되는 하늘에 집어 넣습니다.
- 파이프는 그냥 공중에 떠 있는 것이 아니라 반드시 딱딱한 블럭들 위에 있어야 한다는 것을 알고 있습니다.
- 적절한 장소에 적을 배치합니다.
- 플레이어가 앞으로 나아갈 수 없도록 막는 어떠한 것도 만들지 않습니다.
- 게임에 들어 있는 실제 레벨에서 그 스타일을 따왔기 때문에, 슈퍼 마리오 브라더스 1의 실제 레벨 같이 느껴집니다.
자 이제, 이 레벨을 가져와 Super Mario Maker에서 재현해 보겠습니다:

이제 직접 플레이 해보세요!
Super Mario Maker를 사용하는 경우 온라인으로 북마크하거나 레벨 코드 4AC9–0000–0157-F3C3을 사용하여 찾아보면 이 레벨을 직접 플레이 할 수 있습니다.
장난감 vs. 실제 애플리케이션
우리가 모델을 훈련시키는데 사용한 순환 신경망(recurrent neural network 또는 RNN) 알고리즘은 실제 회사에서 음성 인식이나 번역과 같은 어려운 문제를 해결하는 데 사용하는 알고리즘과 같은 종류입니다. 우리 모델이 최첨단이 아닌 ‘장난감’으로 보이는 이유는 단지 매우 적은 데이터로 생성되었기 때문입니다. 초기 슈퍼 마리오 브라더스 게임은 정말 좋은 모델을 만들기 위한 충분한 데이터를 제공할 만큼 많은 레벨이 없을 뿐입니다.
닌텐도가 보유하고 있는 수십만 개의 사용자 제작 Super Mario Maker 레벨에 접근할 수 있다면, 우리는 정말 놀라운 모델을 만들 수도 있습니다. 그러나 닌텐도가 이를 제공하지 않기 대문에 불가능합니다. 대기업들은 데이터를 무료로 제공하지 않습니다.
더 많은 산업 분야에서 기계 학습이 더욱 중요해 짐에 따라, 좋은 프로그램과 나쁜 프로그램의 차이는 모델을 훈련시키는 데 얼마나 많은 양의 데이터를 확보했느냐가 될 것입니다. 그렇기 때문에 Google이나 Facebook과 같은 회사들이 그렇게도 여러분의 데이터를 좋아하는 것입니다.
예를 들어, Google은 대규모 기계 학습 애플리케이션을 구축하기 위한 소프트웨어 툴킷인 TensorFlow를 오픈소스로 공개했습니다. Google이 이처럼 중요하고 뛰어난 기술을 무료로 제공 한 것은 꽤나 큰 사건이었습니다. 이는 Google Translate을 강력하게 하기 위해 이를 공개했던 이유와 같습니다.
*역자주: 오픈소스를 통해 많은 사람이 사용하면 수많은 훈련 데이터가 축적되어 그 프로그램의 성능이 당연히 강력해지기 때문입니다.
즉, Google이 수집한 모든 언어에 대한 대규모 데이터가 없다면, 당신은 절대 Google Translate에 대한 경쟁 제품을 만들 수 없습니다. 데이터야 말로 Google의 가장 큰 장점입니다. 나중에 Google 지도 위치 기록(Google Maps Location History) 또는 Facebook 위치 기록(Facebook Location History)을 열어보면 당신이 이전에 가봤던 모든 장소가 저장되어 있다는걸 알게 될 것입니다.
추가 읽을 거리
기계 학습에서 문제를 해결할 수있는 유일한 방법은 없습니다. 어떻게 데이터를 사전 처리할지 또는 어떤 알고리즘을 사용할지 결정할 때 우리에겐 무한한 옵션이 있습니다. 때때로 여러 접근방법을 결합하면 하나의 접근 방법보다 더 나은 결과를 얻을 수 있습니다.
이 글을 읽은 분들이 슈퍼 마리오 레벨을 생성하는 또다른 흥미로운 접근 방법에 대한 링크들을 보내왔습니다.
- Amy K. Hoover 팀은 레벨의 객체(파이프, 바닥, 플랫폼 등) 타입을 전체 교향곡속에서 하나의 목소리으로 나타내는 접근 방식을 사용했습니다. Functional Scaffolding이라는 프로세스를 사용해서, 시스템은 주어진 객체 타입의 블록으로 레벨을 보강해 줄 수 있습니다. 예를 들어, 당신이 레벨의 기본 모양을 스케치 하면, 시스템이 파이프 및 질문 블록을 추가하여 디자인을 완성해 줍니다.
- Steve Dahlskog 팀은 레벨 데이터의 각 열을 일련의 n-gram “글자들(words)”로 모델링하면 대형 RNN보다 훨씬 간단한 알고리즘으로 레벨을 생성 할 수 있다는 것을 보여주었습니다.
'Machine Learning' 카테고리의 다른 글
| #5. Training / Test / Validation Set : 오버피팅을 피하는 방법 Terry TaeWoong Um (0) | 2016.12.27 |
|---|---|
| Machine Learning Matlab lectures (0) | 2016.12.20 |
| Machine Learning & Deep Learning Tutorials (0) | 2016.12.19 |
| Jupyter notebook content for my OReilly book, the Python Data Science Handbook (0) | 2016.12.17 |
| This AI-augmented microscope uses deep learning to take on cancer (0) | 2016.12.17 |